EnzymoGenius™ leads in the field of bioinformatics-driven enzyme active site prediction, employing cutting-edge tools to unravel the intricacies of enzyme functionality. Our service is dedicated to providing precise insights into the location and characteristics of active sites within enzymes, facilitating a deeper understanding of their catalytic capabilities.
Overview
Bioinformatics tools play a pivotal role in the accurate prediction of enzyme active sites, a critical aspect in understanding catalytic mechanisms and designing targeted inhibitors. Leveraging computational algorithms and structural bioinformatics, researchers have made substantial progress in elucidating enzyme active site features. Through the analysis of protein structures and sequences, these tools facilitate the identification of key residues and motifs governing enzymatic activity. Molecular docking simulations further refine predictions, enabling the exploration of substrate binding interactions. The integration of diverse datasets, including genomics and structural databases, enhances the reliability of predictions. Ongoing research focuses on refining algorithms, incorporating machine learning approaches, and adapting to evolving structural biology datasets, fostering continual advancements in enzyme active site prediction for applications in drug discovery and biocatalysis.
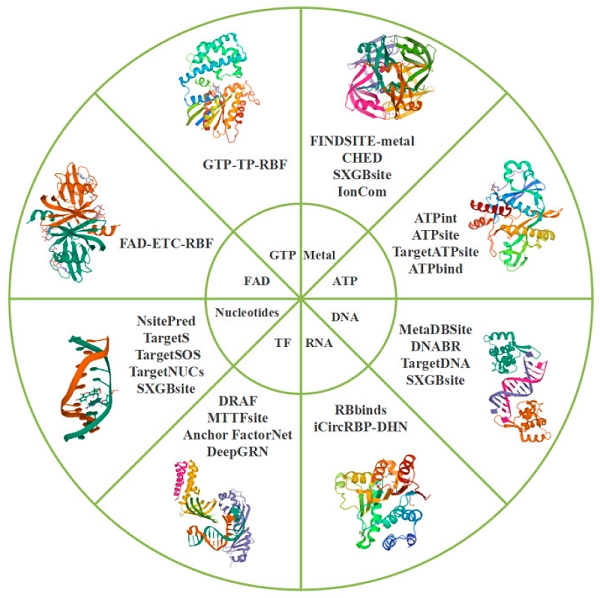 Fig. 1 Tools used in ligand-specific methods to predict binding sites. (Liao J, et al., 2022)
Fig. 1 Tools used in ligand-specific methods to predict binding sites. (Liao J, et al., 2022)
Our Services
- Sequence Analysis
Leveraging bioinformatics algorithms to analyze enzyme sequences and identify conserved motifs associated with active sites.
- Structural Bioinformatics
Utilizing three-dimensional structural information to predict potential active site locations and their structural features.
- Functional Residue Prediction
Identifying key amino acid residues crucial for enzyme activity through advanced computational analyses.
- Phylogenetic Profiling
Assessing evolutionary relationships to enhance the prediction accuracy of enzyme active sites across related species.
Key Differentiators From Other Companies
- Integrated Approach
EnzymoGenius™ employs a holistic methodology that combines sequence, structural, and evolutionary information for comprehensive active site predictions.
- Machine Learning Integration
Our service incorporates machine learning algorithms to enhance prediction accuracy and adapt to evolving bioinformatics methodologies.
- Validation Protocols
Rigorous validation processes ensure the reliability and accuracy of predicted enzyme active sites, setting our service apart in terms of precision and trustworthiness.
Application Areas We Can Serve
- Drug Target Identification
Facilitating the discovery of potential drug targets by pinpointing active sites essential for enzyme function.
- Enzyme Engineering
Guiding the modification and optimization of enzymes for industrial applications through targeted active site predictions.
- Bioprocess Optimization
Enhancing the efficiency of bioprocesses by identifying and manipulating key active sites in enzymes involved in bioconversion.
- Functional Annotation
Contributing to the functional annotation of enzymes in genomic studies by elucidating their active site characteristics.
EnzymoGenius™ stands at the forefront of bioinformatics-driven enzyme active site prediction, offering a comprehensive service that integrates sequence analysis, structural bioinformatics, functional residue prediction, and phylogenetic profiling. What sets us apart is our integrated approach, incorporation of machine learning, and adherence to rigorous validation protocols, ensuring unparalleled accuracy and reliability. Contact us to unlock the potential of bioinformatics in predicting enzyme active sites.
Reference
- Liao, J.; et al. In Silico Methods for Identification of Potential Active Sites of Therapeutic Targets. Molecules. 2022, 27(20): 7103.

































 Fig. 1 Tools used in ligand-specific methods to predict binding sites. (Liao J, et al., 2022)
Fig. 1 Tools used in ligand-specific methods to predict binding sites. (Liao J, et al., 2022)