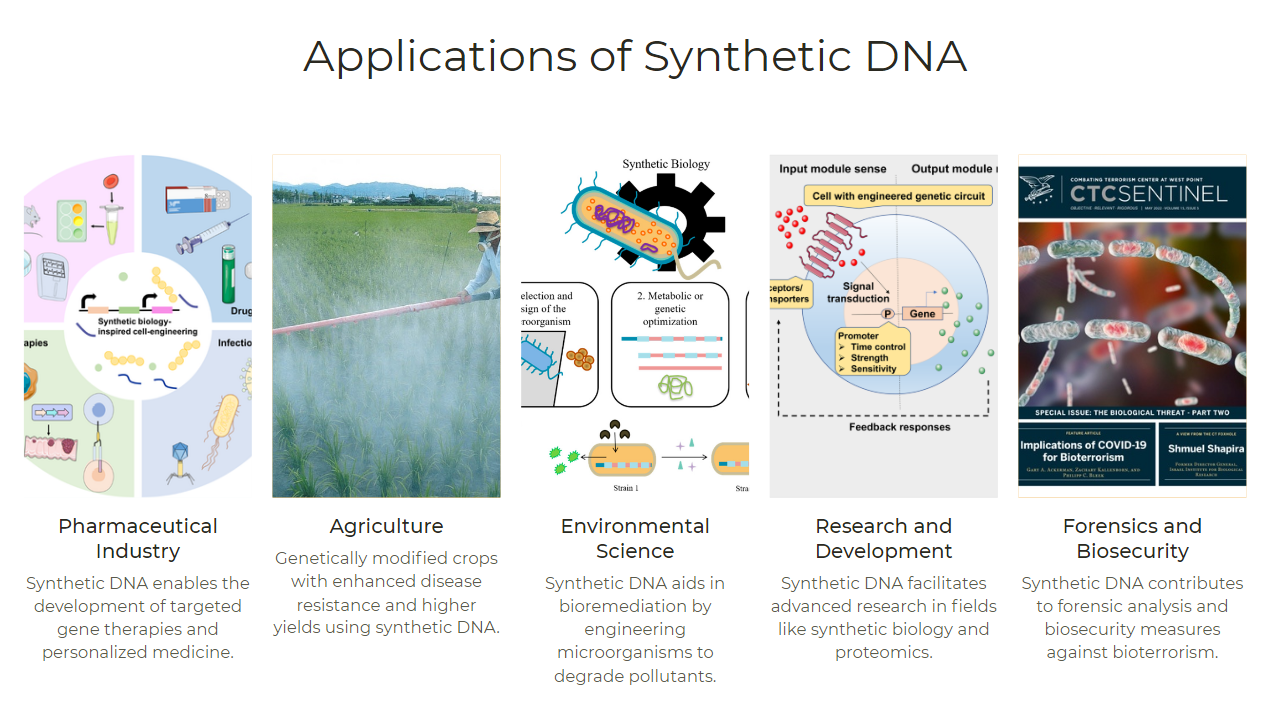
DNA is a fundamental component of life and a core material in synthetic biology. Synthetic biology is an emerging interdisciplinary field that utilizes engineering principles and techniques to design and construct biological systems with specific functions. The application prospects of synthetic biology are very broad, involving multiple fields such as medicine, agriculture, energy, environment, and information. For example, through synthetic biology, we can develop new vaccines and drugs, modify microorganisms to produce renewable energy and chemicals, and even create new artificial life.
To achieve these goals, we need to be able to synthesize DNA quickly, accurately, and at low cost. DNA is a long chain molecule composed of four types of bases (A, T, C, G), carrying genetic information and instructions. By altering the DNA sequence, we can alter the traits and behavior of living organisms. Therefore, customizing DNA is the cornerstone of synthetic biology.
Related Services
However, current DNA synthesis techniques still face many challenges and limitations. Traditional chemical methods are unable to efficiently synthesize long or complex sequences of DNA, and can result in toxic waste and errors. To overcome these issues, some companies and research institutions are developing a new generation of enzymatic DNA synthesis technology that utilizes enzymes present in nature to mimic the DNA synthesis process within cells. Enzymatic DNA synthesis technology has many advantages, such as high fidelity, low toxicity, high efficiency, and low cost. Enzymatic DNA synthesis technology can also enable the development of desktop DNA printers, allowing laboratories to generate the required DNA on demand internally.
Recently, Caroline Seydel published an article titled “DNA writing technologies moving towards synthetic genes” in Nature on October 26, 2023. Introduced the latest progress and future prospects of enzymatic DNA synthesis technology. The article points out that enzymatic DNA synthesis technology may accelerate the pursuit of creating a fully synthetic genome, which was once unimaginable but now seems feasible. The fully synthetic genome refers to the complete genetic material designed and constructed from scratch, which can endow organisms with new functions or characteristics. For example, scientists have successfully synthesized yeast chromosome 2 and bacterial genome 3 from scratch, and are attempting to synthesize a more complex eukaryotic genome 4.

The principle and application of enzymatic DNA synthesis technology
Enzymatic DNA synthesis technology mainly utilizes an enzyme called terminal deoxyribonucleotide transferase (TdT) to construct DNA sequences from scratch. TdT is an enzyme that can randomly add bases to the end of a DNA strand without a template. By connecting TdT with specific bases and controlling its interaction with DNA primers fixed on solid supports, bases can be added one by one in a predetermined order and the DNA strand can be gradually extended (Figure 1). After adding each base, it is necessary to clean the excess reactants and remove the bases connected to TdT for the next cycle.

Figure 1: Synthesis and modification of high molecular weight DNA using terminal deoxyribonucleotide transferase. Figure 1 is adapted from Yoo, E., Choe, D., Shin, J., Cho, S.&Cho, B.-Y. Compute Structure Biotech J. 19, 2468-2476 (2021), CC BY 4.0
Using this technology, some companies have successfully synthesized oligonucleotide 5 with over 1000 bases and are continuously increasing production and quality. Enzymatic DNA synthesis technology can not only synthesize long stranded DNA, but also synthesize complex sequences that are difficult to manufacture chemically, such as highly repetitive or GC rich sequences. These sequences have important functions in biology, such as regulating gene expression or forming special structures. In addition, enzymatic DNA synthesis technology can also be carried out in aqueous solutions, avoiding the use of toxic organic solvents and chemicals, thereby reducing environmental pollution and safety risks.
An important application of enzymatic DNA synthesis technology is the development of desktop DNA printers. A desktop DNA printer is an instrument that can generate the required DNA inside the laboratory, including software, reagent kits, and hardware components. Users only need to input the desired DNA sequence to obtain the desired oligonucleotides overnight. In this way, users do not need to wait for delivery from third-party companies, nor do they need to worry about supply chain disruptions or delays. Desktop DNA printers can also enhance user control and confidentiality of their data, avoiding the leakage of sensitive or proprietary information.
Gene Assembly Tools
| Catalog Number | Product Name | Product Size | Applications | Price |
|---|---|---|---|---|
| GE0011 | High-Fidelity DNA Assembly Cloning Kit | 10 Reactions | Function as a DNA-guided endonuclease. | Online Inquiry |
| GE0012S | High-Fidelity DNA Assembly Master Mix | 10 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |
| GE0012L | High-Fidelity DNA Assembly Master Mix | 50 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |
| GE0012X | High-Fidelity DNA Assembly Master Mix | 250 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |
The advantages and challenges of desktop DNA printers
Desktop DNA printers have great appeal for laboratories of various sizes and fields. It can accelerate the design build test learn cycle within the laboratory, improve productivity and innovation capabilities. For example, in fields such as virology, diagnostics, and drug development, laboratories need to quickly generate various variants or customized oligonucleotides for testing or validation. If the required oligonucleotides can be printed internally, it can greatly shorten the experimental cycle and respond promptly to emerging epidemics or challenges.
Desktop DNA printers can also reduce laboratory operating costs and waste. Due to the desktop DNA printer’s ability to generate the required amount of oligonucleotides on demand, laboratories do not need to bulk order excess oligonucleotides to save on shipping costs, nor do they need to invest additional resources in storing and managing these oligonucleotides. In addition, due to the use of water-soluble reagents and the minimal waste generated by desktop DNA printers, laboratories can also reduce environmental impact and save on the cost of handling toxic chemicals.
Of course, desktop DNA printers also face some challenges and limitations. Firstly, desktop DNA printers are currently unable to synthesize oligonucleotides with more than 200 bases, which means users still need to use modular assembly techniques to construct longer genes or fragments. Secondly, the cost of each base in desktop DNA printers may be higher than that of large-scale suppliers of oligonucleotides, which may not be an economical choice for users who require large or low-cost oligonucleotides. Thirdly, the use of desktop DNA printers also requires adherence to certain ethical and legal norms to prevent the misuse or misuse of DNA, resulting in biosafety or social risks.
Therefore, a hybrid market model may emerge in the future, allowing users to choose the most suitable DNA synthesis method based on their own needs and conditions. Desktop DNA printers and large-scale suppliers both have their own advantages and disadvantages, and they can complement or compete to jointly promote the development and innovation of DNA synthesis technology.
In summary, enzymatic DNA synthesis technology is a breakthrough technology that provides a powerful and sustainable tool for synthetic biology. It enables us to create new DNA sequences with unprecedented speed and accuracy, and apply them to various fields. With the popularization and improvement of desktop DNA printers, we can expect to see more amazing and beneficial innovations and discoveries.