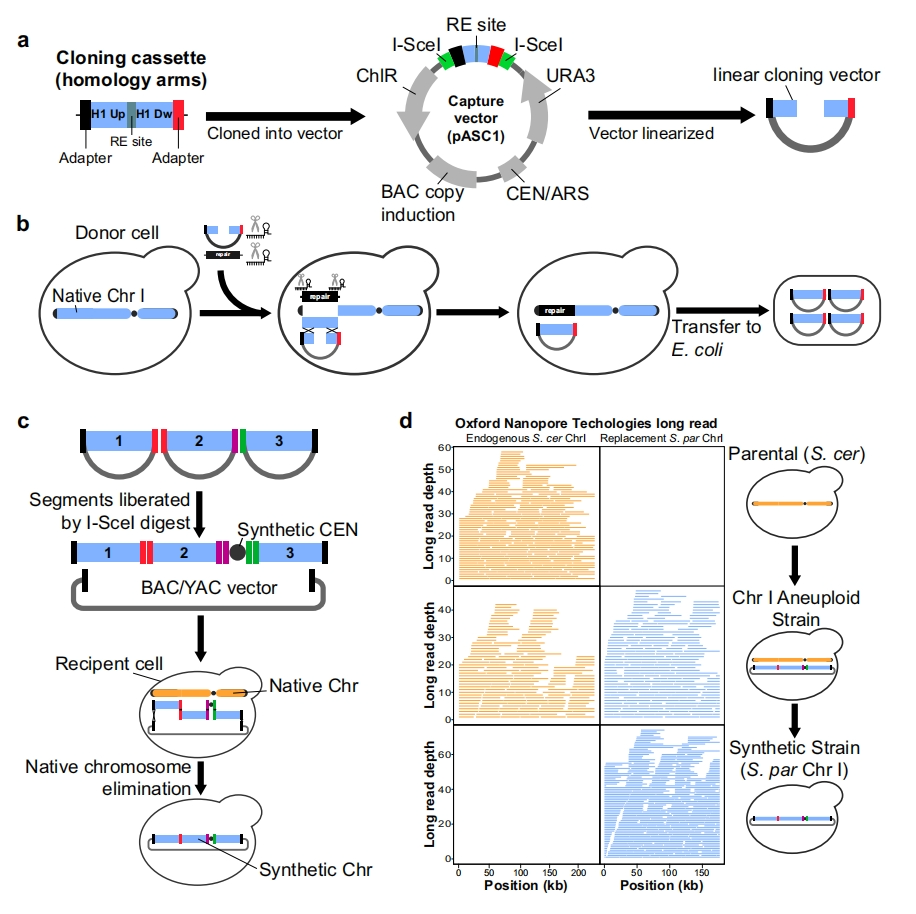
Evolution has produced multiple functional proteins that can precisely regulate cellular processes. In recent years, deep generation models have emerged, aiming to learn from this diversity and generate both effective and novel proteins, with the ultimate goal of customizing functionality to address today’s prominent challenges.
Evolution has produced multiple functional proteins that can precisely regulate cellular processes. In recent years, deep generation models have emerged, aiming to learn from this diversity and generate both effective and novel proteins, with the ultimate goal of customizing functionality to address today’s prominent challenges.
When it comes to creating new proteins on computers, deep generation models are becoming increasingly powerful tools. The diffusion model is a type of generative model that has recently been proven to generate physiologically reasonable proteins. Unlike any actual protein seen in nature, it can provide unparalleled ability and control in de novo protein design.
However, the current state-of-the-art models for constructing protein structures severely limit the breadth of their training data and limit them to small and biased parts of the protein design space.
Microsoft researchers have developed EvoDiff, a universal diffusion framework that combines evolutionary scale data with the unique regulatory capabilities of diffusion models to create adjustable proteins in sequence space. EvoDiff can diversify structurally reasonable proteins, covering all possible sequences and functions. EvoDiff can construct proteins that cannot be accessed by structure based models, such as those with disordered parts, while also designing scaffolds for useful structural motifs, which proves the universality of sequence based formulas.
In protein sequence evolution, EvoDiff is the first deep learning framework to demonstrate the effectiveness of diffusion generated models.
Ava Amini, co author of EvoDiff and senior researcher at Microsoft, said, “If there is anything we can learn from EvoDiff, I believe it is that we can and should generate proteins through sequences because we can achieve universality, scale, and modularity. Our diffusion framework gives us the ability to do this and controls how we design these proteins to meet specific functional goals
Kevin K. Yang, another co author of EvoDiff, stated, “We envision that EvoDiff will expand the capabilities of protein engineering beyond the structure function paradigm to shift towards programmable, sequence first design. Through EvoDiff, we demonstrate that we may not actually need structure, but rather ‘protein sequences are what you need’ to design new proteins in a controllable manner
This study is titled ‘Protein generation with evolutionary diffusion: sequence is all you need’ and published on the bioRxiv preprint platform.

GitHub link: https://github.com/microsoft/evodiff
Paper link: https://doi.org/10.1101/2023.09.11.556673
640 million parameters
The core of the EvoDiff framework is a model with 640 million parameters, which is trained based on data from all different species and protein functional categories. The data for the training model comes from the OpenFold dataset UniRef50 used for sequence alignment, a subset of UniProt data, and the protein sequence and functional information database maintained by the UniProt alliance.
Uniref50 is a dataset containing approximately 42 million protein sequences. The MSA comes from the OpenFold dataset, which includes 16000000 UniCluster30 clusters and 401381 MSAs, covering 140000 different PDB chains. The information about IDR comes from reverse homologous GitHub.
The main features of EvoDiff
The main features of EvoDiff are as follows:
- In order to generate manageable protein sequences, EvoDiff combines evolutionary scale data with diffusion models.
- EvoDiff can diversify structurally reasonable proteins, covering all possible sequences and functions.
- In addition to generating proteins with disordered parts and other features that cannot be obtained by structure based models, EvoDiff can also generate scaffolds for functional structural motifs, demonstrating the universal applicability of sequence based formulas.
EvoDiff is a novel generative modeling system developed by combining evolutionary scale datasets with diffusion models for creating programmable proteins solely from sequence data. It uses a discrete diffusion framework, where the forward process iteratively disrupts the protein sequence by changing its amino acid properties, and the neural network parameterized learning reverse process utilizes a natural framework to predict the changes made at each iteration. Proteins serve as sequences of discrete markers in amino acid language.

Figure 1: EvoDiff, used for controllable protein design only from sequence data. (Source: Paper)
Reverse methods can be used to create protein sequences from scratch. Compared with the traditional continuous diffusion formula used in protein structure design, the discrete diffusion formula used in EvoDiff has made significant mathematical improvements. Multiple sequence alignment (MSA) highlights the conserved patterns and variations of amino acid sequences in relevant proteomes, thereby capturing evolutionary connections beyond the evolutionary scale dataset of individual protein sequences. In order to utilize this additional depth of evolutionary information, they constructed a discrete diffusion model trained on MSA to generate novel single lines.
Creating adjustable proteins in sequence space
To demonstrate its effectiveness in tunable protein design, researchers examined sequences and MSA models (EvoDiff Seq and EvoDiff MSA, respectively) on a series of spectra generating activities.
They first demonstrated that EvoDiff Seq can reliably produce high-quality and diverse proteins, accurately reflecting the composition and function of natural proteins. EvoDiff-MSA guides the development of new sequences by comparing proteins with similar but unique evolutionary histories. Finally, they demonstrated that EvoDiff can reliably generate proteins with IDR, directly overcoming the key limitations of structure based generation models, and can successfully generate scaffolds with functional structural motifs without any clear structural information by utilizing the regulatory function of diffusion based modeling frameworks.

Figure 2: EvoDiff-MSA supports evolutionary guided sequence generation. (Source: Paper)
In order to generate diverse new proteins with the possibility of regulation based on sequence constraints, researchers proposed EvoDiff, a diffusion modeling framework. By challenging the structure based protein design paradigm, EvoDiff can unconditionally sample structurally reasonable protein diversity by generating essentially disordered regions and scaffold structural motifs from sequence data.
By guiding and adjusting, the created sequence can be iteratively adjusted to meet the required quality, which can be added to these functions in future research. The EvoDiff-D3PM framework is suitable for conditional adjustment through guidance, as the identity of each residue in the sequence can be edited at each decoding step.
However, researchers have observed that OADM typically outperforms D3PM in unconditional generation, possibly because OADM denoising tasks are easier to learn than D3PM. Unfortunately, OADM and other existing conditional LRAR models such as ProGen have reduced the effectiveness of guidance. It is expected that new protein sequences will be generated by adjusting the functional targets of EvoDiff-D3PM, such as those described by sequence function classifiers.
EvoDiff data requirements are extremely low
EvoDiff has extremely low data requirements, which means it can easily adapt to subsequent uses, which can only be achieved through structure based methods. Researchers have shown that EvoDiff can create IDRs through repair without the need for fine-tuning, thus avoiding the classic traps of structure based prediction and generation models.

Figure 3: EvoDiff generates essentially unordered regions. (Source: Paper)
The high cost of obtaining a large sequencing dataset structure may prevent researchers from using new biological, medical, or scientific design options that can be unlocked by fine-tuning EvoDiff on application specific datasets such as those from display libraries or large screens. Although AlphaFold and related algorithms can predict the structure of many sequences, they encounter difficulties in point mutations and may be overly confident in indicating the structure of false proteins.
Next step plan
In summary, Microsoft scientists have released a set of discrete diffusion models that can be used for sequence based protein engineering and design. The EvoDiff model can be extended for guided design based on structure or function, and they can be immediately used for unconditional, evolutionary guided, and conditional creation of protein sequences. They hope that by directly using protein language to read and write processes, EvoDiff will open up new possibilities for programmable protein creation.
This is just a model with 640 million parameters, and if we expand to billions of parameters, we may see an improvement in generation quality, “Alamdari said. Although we have demonstrated some coarse-grained strategies, in order to achieve finer grained control, we hope to adjust EvoDiff based on text, chemical information, or other means to specify the required functionality
Next, the EvoDiff team plans to test the proteins generated by the model in the laboratory to determine if they are feasible. If this proves to be the case, they will start developing the next generation framework.