p>The rapid development of synthetic biology is inseparable from the progress of underlying technology. Since the discovery of the double helix structure of DNA in the second half of the 20th century, the three major technologies of gene sequencing, gene editing, and gene synthesis have made rapid progress; At the same time, with the continuous development of systems biology, protein engineering and other technologies, synthetic biotechnology is also expected to usher in a new leap.
1. Gene sequencing technology
Gene sequencing technology has evolved from the first generation to the third generation, improving the efficiency of gene sequencing while also reducing the cost of gene sequencing. In the early era of first generation sequencing technology, the cost of sequencing the entire human genome required $3 billion. However, in 2019, the cost of sequencing the entire human genome had dropped to $1000, and it is expected that its cost will continue to fall below $100 in the next 10 years. Gene sequencing is the foundation of synthetic biology research and an important means for humans to “open their eyes to the world”. The efficiency and cost of gene sequencing directly determine the development of the industry.

Fig1:The decreasing trend of gene sequencing costs

Fig2:The Trend of Improving Gene Sequencing Efficiency
Generation 1: High accuracy, low throughput (can only measure a DNA fragment of around 1000bp), and high cost.
Second generation: Low accuracy (requires splicing and mismatches), high throughput (can measure multiple 300bp DNA fragments at once), and low cost.
Third generation: High accuracy, high throughput (multiple DNA fragments of over 1000bp can be measured at once), and high cost.
2.Gene editing technology
Gene editing technology determines the targeted modification of the genetic material of living organisms, which can achieve changes in their traits and metabolism. It is a very important fundamental tool in synthetic biology.
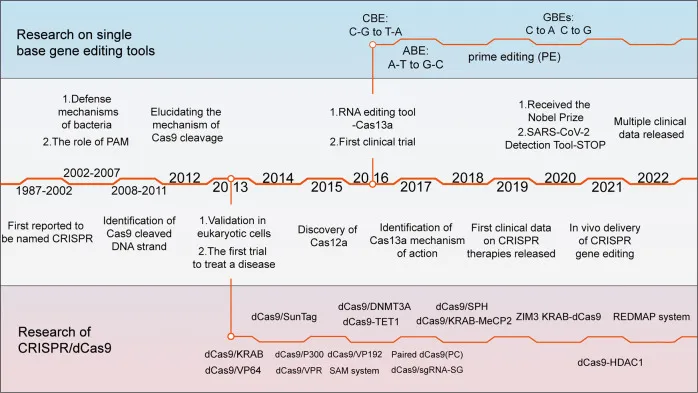
The progress of gene editing technology has gone through the first generation of zinc finger protein technology, the second generation of transcriptional activator like effector (TALEN) technology, and the third generation of CRISPR/Cas9 technology. CRISPR technology overcomes the drawbacks of traditional gene manipulation such as long cycles, low efficiency, and narrow application, greatly improving the convenience of gene manipulation.
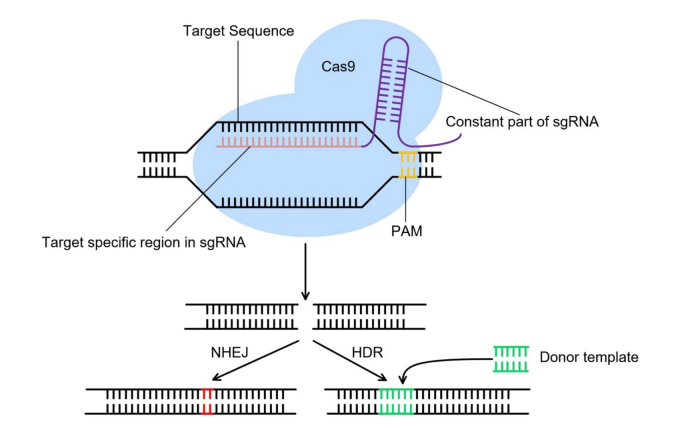
CRISPR is the abbreviation for clusters of regularly spaced short palindromic repeats, and is a part of the bacterial immune system. Therefore, CRISPR was first modified from bacteria. To resist foreign viral infections, bacteria cut and destroy the DNA of invading viruses. When bacteria survive a virus infection, they retain a portion of the virus DNA for recording. When the same virus invades again the next time, the bacteria can quickly recognize the invading virus and destroy the virus DNA. The CRISPR gene editing system mainly consists of two parts: 1) Cas9 protein, which acts as a “scissors”; 2) Customizable RNA guide sequences, specifically DNA sequences, serve as’ guides’. To edit genes, you need to use “scissors” to first remove the unwanted parts. The cutting position needs to be guided by a “guide”. RNA guide sequences are single stranded nucleotides that complement specific splicing target genes. For example, a mutated gene sequence that produces sickle like cells can serve as a target sequence for guiding RNA. The Cas9 protein “reads” the host’s DNA through guided RNA, thereby finding the target gene. When the guide RNA is paired with the target sequence, it usually triggers the cleavage mechanism of the Cas9 protein itself, thereby cleaving the nucleotide bonds of the target gene.

Fig3:CRISPR technology
CRISPR technology has powerful gene editing capabilities and has been widely applied in the editing of microorganisms, animal and plant cells, and has also made progress in the clinical treatment of human genetic diseases. However, it still faces ethical risks and off target effects, among which off target effects can lead to positional risks.
In addition, many gene editing methods suitable for non host use have also emerged this year, such as Gibson Assembly, yeast homologous recombination, and Escherichia coli Red recombination. These methods will integrate DNA fragments from different organisms into engineering hosts, achieving downstream research and applications in metabolic engineering, metabolomics, and heterologous expression.
| Catalog Number | Product Name | Product Size | Applications | Price |
|---|---|---|---|---|
| GE0011 | High-Fidelity DNA Assembly Cloning Kit | 10 Reactions | Function as a DNA-guided endonuclease. | Online Inquiry |
| GE0012S | High-Fidelity DNA Assembly Master Mix | 10 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |
| GE0012L | High-Fidelity DNA Assembly Master Mix | 50 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |
| GE0012X | High-Fidelity DNA Assembly Master Mix | 250 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |
3 Gene sequencing technology
Gene editing technology focuses on re editing local DNA sequences, while gene synthesis technology focuses on the artificial synthesis of large fragments of DNA. DNA is the genetic material of life. The breakthrough in DNA synthesis technology has truly made humanity the ‘creator’.
Oligonucleotide synthesis began in the 1950s. By the 1980s, columnar synthesis technology had been developed, and to this day, automated equipment has gradually matured. However, it still has low efficiency and high cost, making it suitable for the synthesis of short stranded DNA. Other methods are also gradually being researched and applied: in recent years, chip DNA synthesis technology has shown certain cost advantages in long-chain DNA synthesis; The enzymatic synthesis technology is currently in the research stage.
4 Systems Biology Technology
Systems biology is the discipline that studies the composition of all components in a biological system and the interrelationships between these components under specific conditions. That is to say, systems biology is different from previous experimental biology – it only focuses on individual genes and proteins, it studies all genes, all proteins, and all interrelationships between components. The goal of systems biology is to “digitize” the analysis of life activities.

Fig4:Transcriptomics processing large amounts of data
The main technological platforms of systems biology include genomics (DNA), transcriptomics (mRNA), proteomics (proteins/enzymes), metabolomics (products), etc.
Due to the global, systematic, and digital nature of the data collected in systems biology, which generates a large amount of data, manual processing is not feasible. Therefore, the connection between systems biology and computer science, statistics, and other disciplines is very close.
Currently, systems biology is still mainly focused on research, with high costs and a long cycle, so its application is relatively limited; But its potential application value is very high. In the field of synthetic biology, metabolomics is a very suitable method for adjusting the production indicators of target products.
5.Directed evolution techniques for enzymes
Enzymes are catalysts in microorganisms, responsible for the transformation of substances, and their efficiency is closely related to their structure. In theory, any chemical reaction can achieve enzyme catalysis (such as the synthesis of starch from carbon dioxide).
Enzymes are a type of protein composed of amino acid residues and have a quaternary structure, whose three-dimensional structure determines catalytic activity. Changing amino acid residues can achieve changes in enzyme structure, thereby altering catalytic activity. It mainly involves changing the amino acid sequence near the active pocket of the protein, resulting in a change in its affinity for the substrate.

Fig5:Activity Pocket of Enzymes
There are a total of 22 common amino acids, and there are usually several amino acid residues in the active pocket position. Therefore, there are many mutants and the workload of modification is huge. Assuming an enzyme has 3 active amino acids, the number of mutants is 21 * 21 * 21=9261.