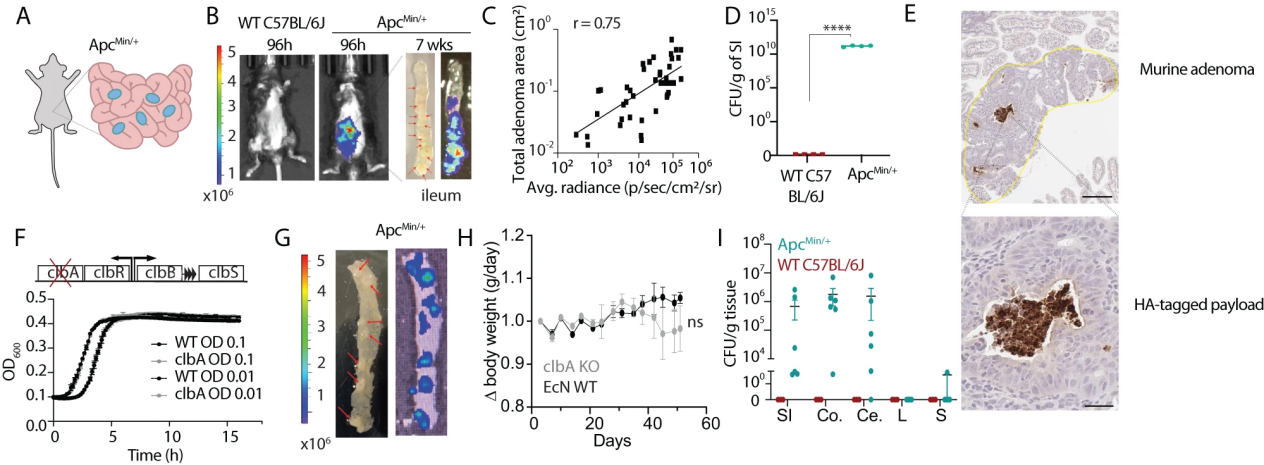
In 2011, countries such as the United States, China, the United Kingdom, Singapore, and Australia launched the “Sc2.0 Project” to redesign and synthesize all 16 chromosomes of brewing yeast. This was the first attempt by humans to design and synthesize the genome of eukaryotes from scratch.
On November 8, 2023, the latest research findings of the Sc2.0 project were published in Cell and its subsidiaries. Among them, Cell Genomics published multiple research findings in cover and album formats. The release of this achievement marks the official completion of the de novo design and synthesis of all chromosomes in the world’s first eukaryotic organism, and significant progress has been made in the scientific milestone project of synthetic biology, laying an important reference for future research in synthetic genomics.

Among them, the research achievements supported by the National Gene Bank Life Big Data Platform were published in Cell Genomics. The research topic is “Dissecting aneuploid phenotypes by constructing Sc2.0 chromosome VII and SCRaMbLEing synthetic disease year”. The de novo design and synthesis of chromosome 7 in brewing yeast were completed, and a non diploid disease model carrying synthetic chromosomes was further constructed, revealing two different pathways behind the recovery of non diploid phenotype in brewing yeast, And discovered genes that play a crucial role in phenotype recovery, providing new strategies for the study of chromosomal abnormalities.

Aneuploidy, usually referring to abnormal chromosome numbers in cells, is the most common variation in the human cancer genome and is closely related to diseases such as embryonic mortality, tumors, and aging [1-3]. Although researchers have widely recognized the important role of aneuploidy in diseases, the significant differences in genetic background, lack of model construction techniques, and the involvement of numerous genes in the DNA regions that form aneuploidy greatly limit the research on the mechanisms and functions related to aneuploidy. As a classic model organism for scientists to study the life activities of eukaryotes, brewing yeast has a relatively simple and representative cell structure and physiological processes, making it one of the ideal model cells for analyzing the complex mechanisms behind aneuploidy.
Related Services
Research background
Unlike the non diploid yeast model based on wild chromosomes constructed in previous studies, an additional chromosome carried in this project is a synthetic type, which contains a large number of recombinase recognition sites, loxPsym, allowing this chromosome to have the ability to induce genome rearrangement (SCRaMbLE technology, one of the core designs of the Sc2.0 project), thereby rapidly generating genetic diversity, Provide a large sample space for downstream scientific and applied research.
In order to obtain this non diploid strain with a clear and highly controllable genetic background, researchers first designed and constructed a synthetic chromosome 7 (synVII) of brewing yeast with a length of 1028 kb, and accurately repaired and characterized it to obtain a synthetic strain that grew similarly to the wild type. Subsequently, a strategy of hindering the separation of target chromosomes was used to construct an additional non whole cell strain (n+synVII) carrying synVII. Through HI-C chromosome capture technology, it was demonstrated that the additional carried synthetic chromosomes were symmetrically arranged with wild chromosomes in the nucleus (Figure 1), and the non diploid strain carrying synthetic chromosomes was more stable than the wild-type non diploid strain.

Figure 1: Strategy for constructing aneuploidy and 3D conformation of chromosomes
Subsequently, based on SCRaMbLE technology, researchers obtained a total of 219 phenotypic recovery strains with different genotypes. Through in-depth analysis of these recovered strains, two different pathways for the restoration of aneuploid phenotypes were discovered: 1. removal of most additional excess chromosomal components; 2. Change the specific region of the target chromosome to correspond to the previous hypotheses of ‘mass action of genes’ and’ few critical genes’ in this field. Interestingly, this study found a strong correlation (p=4.20E-11) between the deletion of a~20 Kb region of synVII and phenotypic recovery through mutation association analysis of partially recovered strains. To further identify the core genes that play a role in this segment, researchers performed stepwise segmentation and single gene knockout validation on the~20 Kb region in the initial non polyploid strain. The experimental results showed that five genes, RPS23A, NUP57, COG2, YGR121W-A, and YGR122W, played a crucial role in phenotype recovery, providing new potential targets for subsequent functional research on non polyploid phenotypes (Figure 2).

Figure 2: Two pathways for the recovery of aneuploid defect phenotype
In summary, this study provides a new strategy for the study of non polyploidy, which can quickly obtain a large number of non polyploid strains with different karyotypes and phenotypes in a short period of time, providing a large amount of research materials for subsequent related studies; On the one hand, this model may have the potential to be applied to the screening of pathogenic targets and related drugs for non polyploid diseases.
Related Products
| Catalog Number | Product Name | Product Size | Applications | Price |
|---|---|---|---|---|
| GE0011 | High-Fidelity DNA Assembly Cloning Kit | 10 Reactions | Function as a DNA-guided endonuclease. | Online Inquiry |
| GE0012S | High-Fidelity DNA Assembly Master Mix | 10 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |
| GE0012L | High-Fidelity DNA Assembly Master Mix | 50 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |
| GE0012X | High-Fidelity DNA Assembly Master Mix | 250 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |