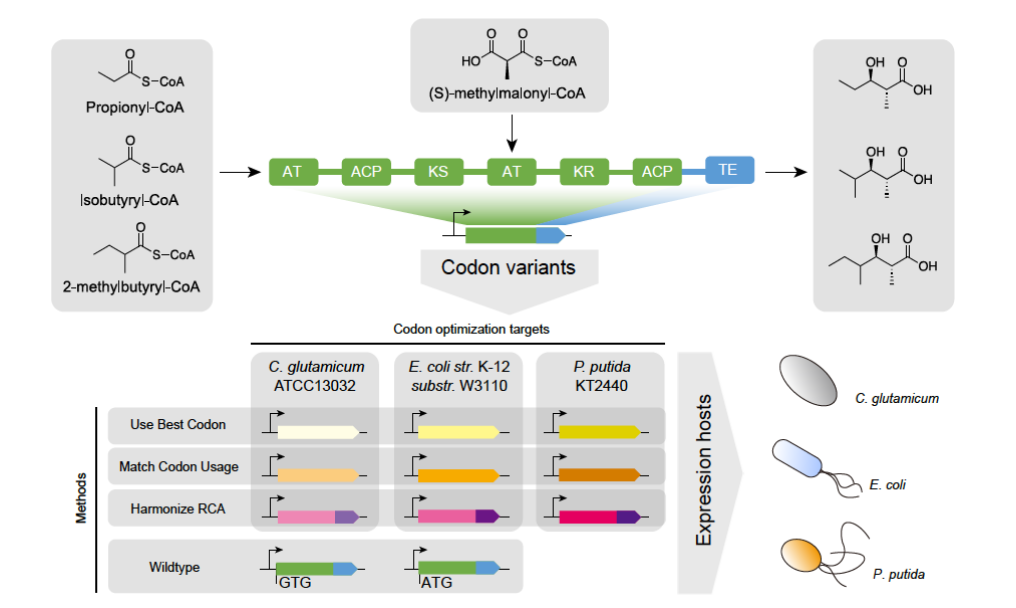
What is miRNA
MiRNA generally refers to micro RNA. MicroRNAs (miRNAs) are a class of non coding single stranded RNA molecules with a length of approximately 19-24nt encoded by endogenous genes, which participate in post transcriptional gene expression regulation in animals and plants.
MiRNAs are usually located between genes or in intronic regions, so it is generally believed that they are not transcribed as part of encoding other types of genes. These small molecule RNAs bind to the 3’UTR region or coding region of the target mRNA sequence through base pairing, cleaving the transcripts of the target genes or inhibiting their translation, thereby regulating gene expression.
CD Biosynsis offers a comprehensive range of RNA synthesis services tailored to meet diverse research needs.
| Service | Description | Applications | Customer Requirements |
|---|---|---|---|
| Custom RNA Synthesis | High-quality RNA molecules synthesized to specific requirements. | Gene expression studies, therapeutic development | Sequence information, desired modifications |
| Custom RNA Oligos Synthesis | Tailored RNA oligonucleotides for various applications. | PCR, qPCR, sequencing | Sequence information, purity requirements |
| Custom siRNA Synthesis | Synthesis of siRNA for gene silencing studies. | Gene knockdown, functional genomics | Target gene sequence, desired modifications |
| Custom miRNA Synthesis | Synthesis of miRNA for microRNA research. | Gene regulation studies, biomarker research | miRNA sequence, specific modifications |
| MicroRNA Agomir/Antagomir Synthesis | Functional studies of microRNA using agomirs and antagomirs. | miRNA function modulation, therapeutic research | Target miRNA sequence, type of modification |
| Long RNA Synthesis Service | Accurate and efficient synthesis of longer RNA sequences. | Vaccine development, long non-coding RNA research | Full sequence information, length requirements |
| Custom RNA Modifications | Specialized RNA modifications for tailored applications. | Structural studies, therapeutic applications | Base sequence, specific modifications |
| Custom sgRNA Libraries | Support for genome editing projects with sgRNA libraries. | CRISPR-Cas9 studies, gene editing research | Target genes, library design specifica |
The formation process of miRNA

At present, people have a relatively clear understanding of the biosynthesis process of animal miRNA. The primary transcript product of miRNA genes in the nucleus, pri miRNA (approximately 300-1000 bases in length), is cleaved by RNase III Drosha enzyme to form pre miRNA (pre miRNA) with a length of approximately 70-90 bases. After preliminary cutting, Pre miRNA is transported from the nucleus to the cytoplasm under the action of the transport protein esportin-5, and then further cleaved by another RNase III – Dicer enzyme to produce mature miRNA (mature miRNA), which is about 20-24 nt long. Mature miRNAs, together with other proteins, form the RISC (RNA induced silencing complex) complex, leading to target mRNA degradation or translation inhibition
Pre miRNA has a hairpin structure but does not have a continuous double stranded structure.
Mature miRNAs can be produced in the 5 ‘and 3’ arms of hair clips, but the frequency of the latter is higher than that of the former.
In plants, the transcription and processing of genes encoding miRNA in the nucleus are coupled, meaning that the formation of miRNA is completed in the nucleus, and there is no transport of miRNA precursors from the nucleus to the cytoplasm. Firstly, the genes encoding miRNA in the nucleus are transcribed under the action of RNA polymerase II to form a primary transcript with a length of about a few hundred nucleotides – pri miRNA; Then, under the action of a type of Dicer enzyme – DCL1, a miRNA prerequisite pre miRNA is formed, which is generally 64-303nt in length, DCL1 continues to act on pre miRNA to form double stranded miRNA; Finally, the double stranded miRNA undergoes methylation modification of the last nucleotide at the 3 ‘end under the action of miRNA methyltransferase HEN1. The main function of methylation is to prevent the activity of transferases and polymerase. The above processes are all completed in the nucleus. Mature miRNAs either bind to ribosomal proteins similar to RISC in the nucleus to form miRNP, which is then transported to the cytoplasm by the homolog of exportin5- HASTY, or are first transported to the cytoplasm by HASTY and then bound to ribosomal proteins to form miRNP.
Transcription and transcription initial product splicing and processing of miRNA coding genes in animal nuclei
Currently, we know very little about how the coding genes of miRNA are transcribed into pri miRNA. Research has found that both RNA polymerase II and polymerase III can participate in the transcription of miRNA. The miRNA genes located within the introns of protein genes are undoubtedly transcribed by polymerase II. Although most other miRNA genes lack signals such as the polyA tail, there is a lot of indirect evidence to suggest that they may also be transcripts of polymerase II, mainly including the following aspects:
Some pri miRNAs are quite long, sometimes exceeding 1kb, which is much longer than the transcripts of polymerase III.
The last base of most pri miRNAs is uracil, which is commonly considered a transcript product of early termination of polymerase III.
The expression of many miRNA genes is variable during development, and this expression change is usually common in the transcripts of polymerase II.
By fusing the 5 ‘region of certain miRNA genes with the development reading framework of the reporter gene, the reporter gene can be expressed, indicating that this miRNA gene can be transcribed through polymerase II.
Research on nematode let-7 has shown that, The let-7 pri miRNA has a polyadenylate tail and is indeed produced by polymerase II transcription.
In addition, polymerase III can also participate in the transcription of miRNA genes. For example, by adding the promoter of polymerase III to the miRNA gene and expressing it through polymerase III, these miRNAs can not only be effectively expressed in vivo, but also perform other functions. This indicates that, The processing, synthesis, and biological functions of miRNA are not necessarily related to which polymerase participates in the transcription of its genes.

The first step of cleavage of pri miRNA is completed by Drosha enzyme, which produces a 70nt precursor of the hairpin structure (pre miRNA). The hairpin contains the required processing sequence on both sides. The double stranded structure can be recognized by the nuclear protein DGCR8, DGCR8 can bind with Drosha enzyme to form pri miRNA processing complexes (as shown in the above figure).
Drosha enzyme contains an RNaseIII domain that can cleave and release hair clips at a distance of approximately 11nt from the base, producing pre miRNA RIIIDa cleaves the 3 ‘- strand of pri miRNA, while RIIIDb cleaves the 5’ – strand. In this process, The combination of CTT and RIIIDb is crucial for stabilizing Drosha. This cleavage reaction not only resulted in the mature 3 ‘end of miRNA, but also caused the 3’ end to protrude from the 2nt, which is the characteristic of cleavage by ribonuclease III. And this cleavage reaction does not occur at the base of the hairpin precursor, but at a base 11nt away from the base.
Pre miRNA is exported from the nucleus via exportin-5
Due to the fact that pre miRNAs require Dicer enzymes in the cytoplasm for processing and processing, transporting them out of the nucleus is a necessary and primary step in the process of becoming mature miRNAs. Pre miRNA enters the cytoplasm from the nucleus through a transport mechanism dependent on RanGTP/esportin-5 (Exp5). Esportin-5 can recognize the protruding 2nt at the 3 ‘end of pre miRNA, promote its release from the Drosha complex, bind with it, and ultimately transport it to the cytoplasm for release. This process is regulated by the concentration of RanGT. Within the nucleus, The concentration of RanGT is relatively high, Esportin-5 can promote the release of pre miRNA from the Drosha complex; And due to the presence of, The concentration of RanGTP is relatively low, Exp5 releases pre miRNA to bind with Dicer enzyme for further cleavage. During transportation, The 3 ‘protruding end of miRNA precursor will facilitate its entry into this pathway.
Dicer processing and miRNA maturation
In the cell nucleus of animals, Drosha’s cleavage produces one end (3 ‘end) of mature miRNA. The cleavage at the other end (5 ‘end) is produced by the cleavage of Dicer enzymes (another type of ribonuclease III) in the cytoplasm. Dicer was first discovered in the study of gene silencing caused by small interfering RNA (siRNA), and later found that it also plays an important role in the maturation process of miRNA.
In fruit flies, nematodes, and mammals, Dicer is necessary for the production of siRNA and miRNA precursors during long dsRNA processing to produce mature miRNA.
Dicer’s involvement in the maturation process of miRNA is similar to the process of producing double stranded RNA in RNA interference. Firstly, Esportin-5 carries pre miRNA into the cytoplasm due to the low concentration of RanGTP in the cytoplasm, Esportin-5 subsequently releases pre miRNA. Dicer enzyme recognizes the double stranded portion of pre miRNA, which is phosphorylated at the 5 ‘end of the stem ring base and has a prominent structure at the 3’ end, with high affinity and complementarity. Then the two spirals at the base of the stem ring are unraveled, Dicer cleaves both strands, cutting off the remaining portion of the precursor to produce a 22 nt long 5 ‘phosphorylated double stranded RNA with 2 nt protrusions at the 3’ end, resembling siRNA with incomplete pairing, known as miRNA: miRNA *. This RNA double strand is composed of complementary strands of mature miRNA and miRNA.
Although Dicer can produce RNA double stranded intermediates during the processing of pre miRNA, these intermediates are usually short-lived and difficult to detect. From the existing mature models of animal miRNAs, the specific cleavage mediated by Drosha initially determines the cleavage site of Dicer in miRNA precursors, which in turn determines the ends of mature miRNAs. Due to research indicating that Drosha has poor efficiency in cleaving non-specific double stranded RNA, while Dicer can cleave any RNA double stranded without sequence dependence, the specificity of miRNA is determined by Drosha rather than Dicer. And the cleavage of Drosha also depends on the secondary structure at the base of the pri miRNA stem ring, as well as the sequence within 125 nt of the outer side of the stem ring base. Dicer cutting requires ATP, ATP may be hydrolyzed by the N-terminal helicase domain. And in miRNA: miRNA * double stranded, only one single strand binds to the RNA induced silencing complex RISC, which then complements the target mRNA, After release, miRNA * is degraded.