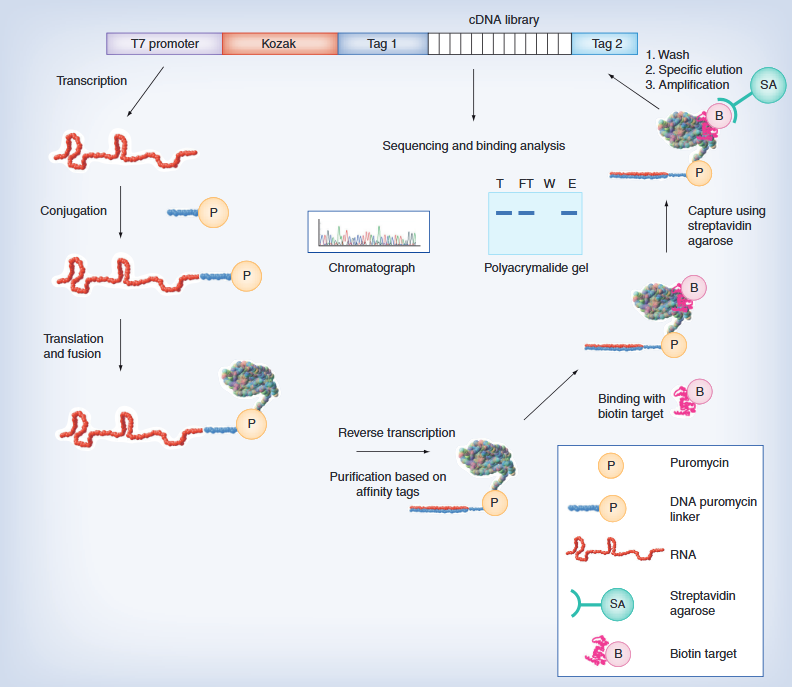
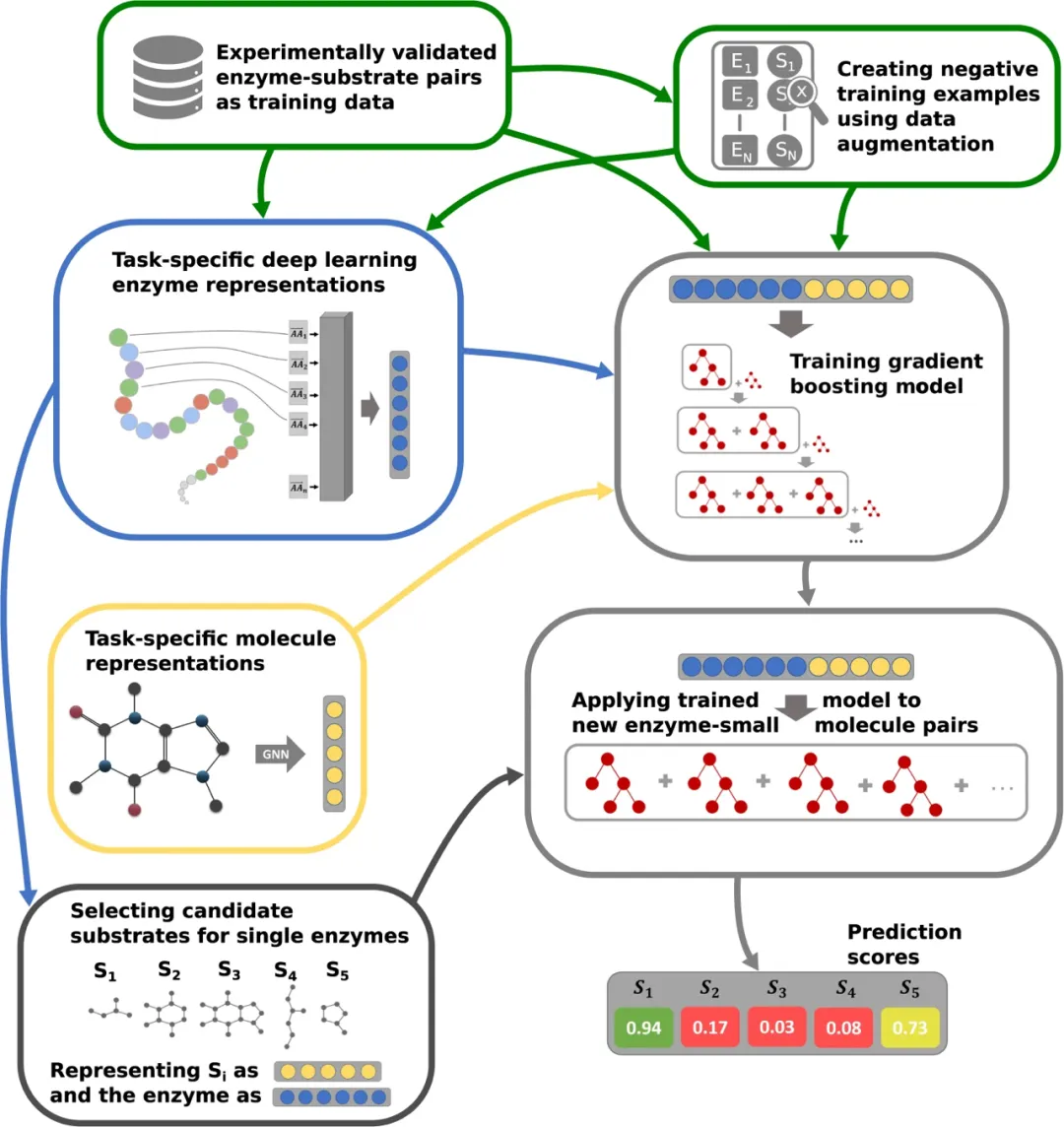
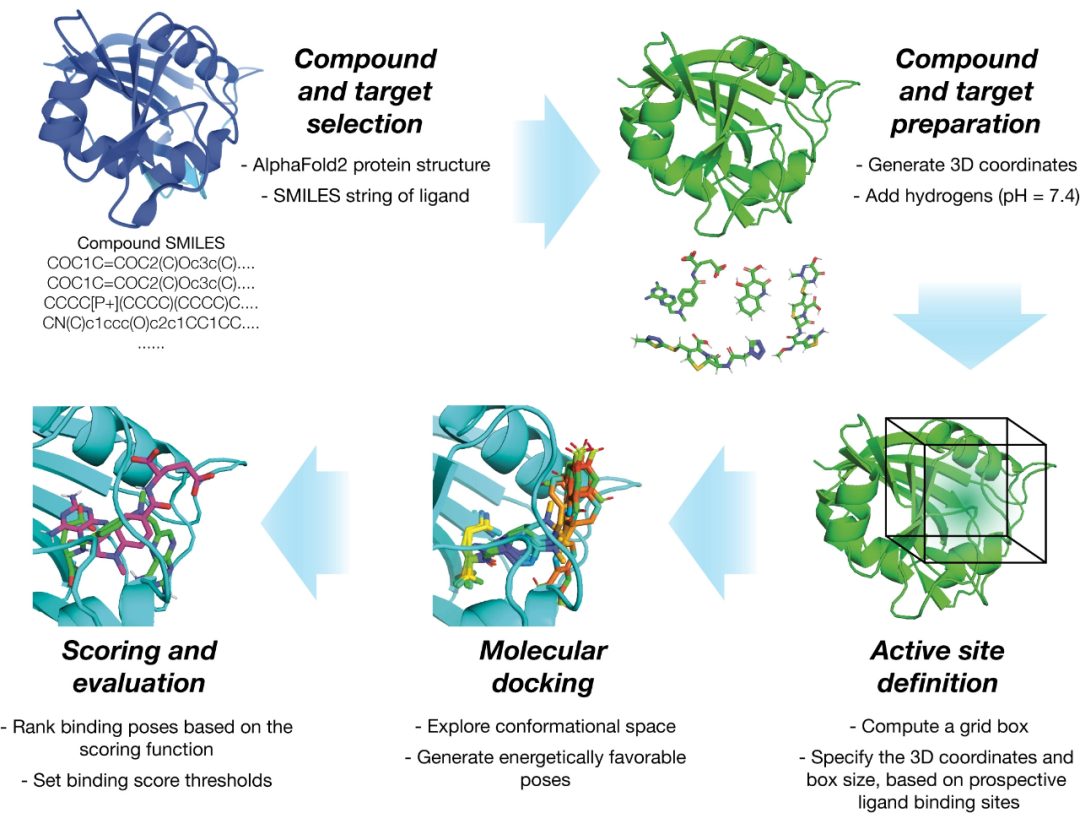
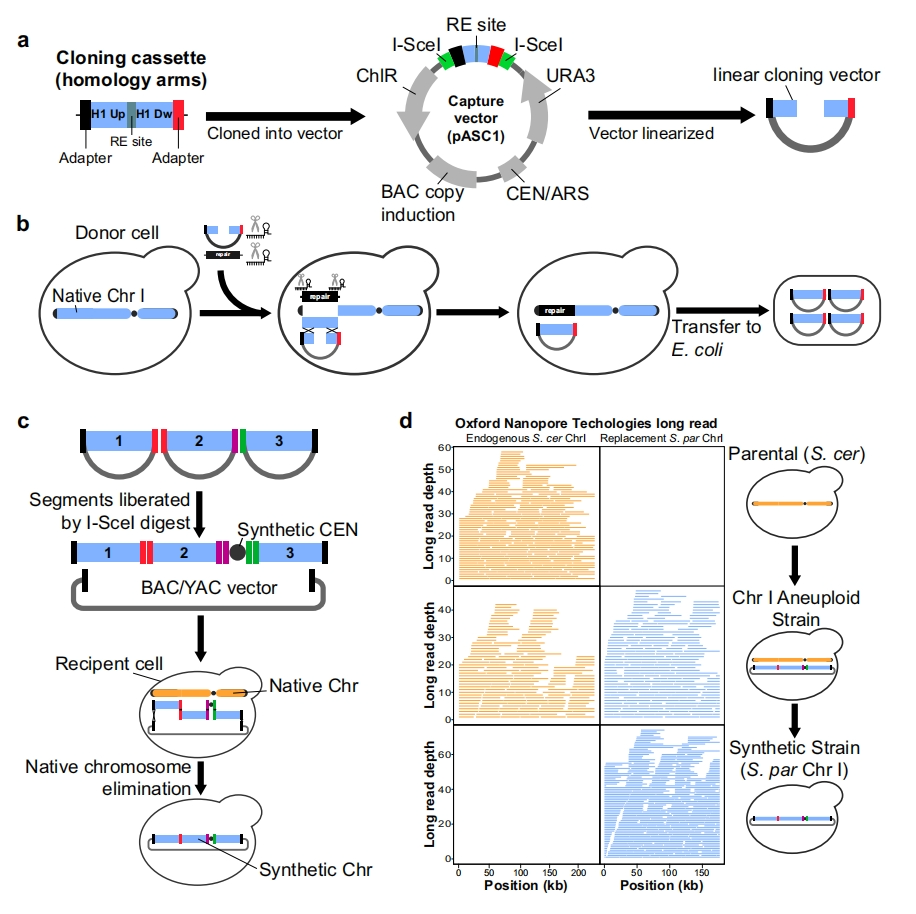
According to a new study by researchers at the University of Michigan, a protein engineering method using simple, cost-effective experiments and machine learning models can predict which proteins are effective for specific purposes.

This method has profound potential in assembling proteins and peptides, and can be used for applications ranging from industrial tools to therapeutic methods. For example, this technology can help accelerate the development of stable peptides to treat diseases in ways that current drugs cannot, including improving the exclusive binding of antibodies to targets in immunotherapy.
“The rules that control how proteins work, from sequence to structure to function, are so complex. Contributing to the interpretability of protein engineering is particularly exciting,” said Marshall Keys, the lead author of this study.
| Catalog Number | Product Name | Product Size | Applications | Price |
|---|---|---|---|---|
| GE0017S | 2-Hydroxy-dATP | 10 μl | Cleanup of sgRNA and Cas9 mRNA. | Online Inquiry |
| GE0017L | 2-Hydroxy-dATP | 5 × 10 μl | Cleanup of sgRNA and Cas9 mRNA. | Online Inquiry |
| GE0018S | 5-Bromo-dUTP | 50 μl | Function as a programmable DNA endonuclease. | Online Inquiry |
| GE0018L | 5-Bromo-dUTP | 5 × 50 μl | Function as a programmable DNA endonuclease. | Online Inquiry |
| GE0019S | 8-Oxo-dATP | 10 μl | In vitro binding of DNA for visualization and target enrichment. |
Online Inquiry |
| GE0019L | 8-Oxo-dATP | 5 × 10 μl | In vitro binding of DNA for visualization and target enrichment. |
Online Inquiry |
| GE0020S | 8-Oxo-dGTP | 30 μl | In vitro digestion of dsDNA. | Online Inquiry |
| GE0020L | 8-Oxo-dGTP | 5 × 30 μl | In vitro digestion of dsDNA. | Online Inquiry |
| GE0021S | dITP Solution | 1 ml | Function as a programmable DNA endonuclease. | Online Inquiry |
| GE0021L | dITP Solution | 10 ml | Function as a programmable DNA endonuclease. | Online Inquiry |
| GE0022 | DNA Shuffling Kit | 15 Reactions | Rapid synthesis of guide RNA. | Online Inquiry |
| GE0023 | dNTP Random Mutagenesis Kit | 15 Reactions | Generation of microgram quantities of custom sgRNA. | Online Inquiry |
| GE0024S | dPTP | 30 μl | Target-specific DNA nicking. | Online Inquiry |
| GE0024L | dPTP | 5 × 30 μl | Target-specific DNA nicking. | Online Inquiry |
| GE0025 | Error Prone Kit | 15 Reactions | Function as an RNA-guided endonuclease. Ideal for direct introduction of Cas9/sgRNA complexes | Online Inquiry |
Currently, most protein engineering experiments use complex, labor-intensive methods and expensive instruments to obtain very accurate data. The lengthy process limits the amount of data that can be obtained, and complex methods are challenging for learning and execution – this is a trade-off of accuracy.
“Our approach suggests that in many applications, you can avoid these complex methods,” said Case.
The updated method first divides cells into two groups, called binary sorting, based on whether they express the desired features – such as binding to fluorescent molecules – or not. Then, the cells are sequenced to obtain potential DNA codes for the proteins of interest. Then, machine learning algorithms will reduce the noise in sequencing data to identify the best proteins.
Greg Thurber, Associate Professor of Chemical Engineering at the University of Michigan and corresponding author of the paper, said, “Instead of choosing the ‘best book’ from the library, it’s better to read many books and then piece together different pages of different stories to find the best book possible, even if it’s not in your original library.” “I was surprised to see the robustness of this technique using simple binary sorting data.”
This method further enhances its accessibility by using linear machine learning models, which are easier to interpret compared to models with dozens of parameters.
Keith said, “Because we can understand the physical rules of how proteins actually work, we can use linear equations to simulate nonlinear protein behavior and create better drugs.”
This study was conducted at the advanced genomics core, structural biology center, biological mass spectrometry equipment, and proteomics and peptide synthesis core.
Related Services
Protein Engineering and Optimization
Synthetic DNA Library Construction