With the continuous intensification of global climate change, environmental pollution and other issues, “clean energy” has become a hot topic on a global scale. As a renewable energy source, clean energy can not only meet the growing energy needs of human society, but also alleviate the current energy crisis and environmental problems to a certain extent.
In recent years, microbial engineering has become increasingly recognized as a potential choice for cleaner and more efficient production of various chemicals required for modern technology. However, many issues have limited the use of this biotechnology. For example, the low efficiency of raw material conversion and the risk of accidental release of genetically modified microorganisms into the environment. The use of live cells also limits their application, as the living system needs to operate within strict conditions, including temperature, salt concentration, and solvent properties. In addition, toxicity or metabolic burden can also limit the mass production of recombinant products.
Related Services
Construction of Artificial Cell Chassis for Biosynthesis
Cellular free metabolic engineering does not rely on metabolic processes carried out by living cells, but rather utilizes the metabolic activity of in vitro cell lysates; Allowing it to be liberated from the requirements of maintaining cell vitality and growth provides greater design flexibility and wider operating conditions for synthetic metabolic pathways. This method provides several potential benefits, including simple operation, high conversion and productivity, and the ability to prevent microbial release in the environment.
In addition, cell-free metabolic engineering systems provide important advantages that cannot be achieved using live cells, including quantitative and precise evaluation of performance through direct sampling, accelerated efficient cycle iteration of design build test learn, and the ability to use non natural or non biological components.

Figure 1: Design Build Test Learn Cycle
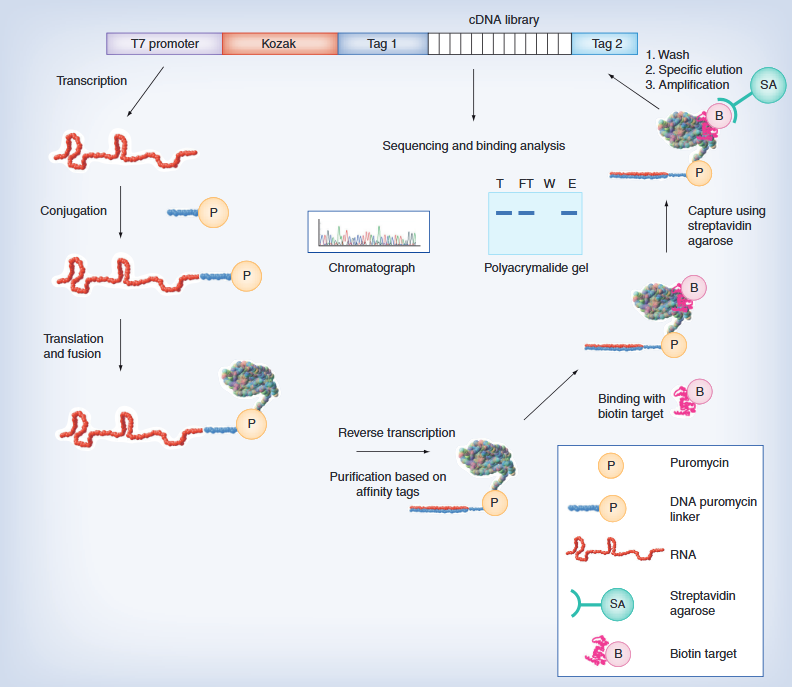
The operational convenience and production capacity of cell-free protein synthesis methods have been continuously developed in the past few decades, and can now directly produce a large number of recombinant proteins from template DNA generated in vitro. Moreover, these advances have successfully combined with the multifunctionality of cell-free protein synthesis, producing enzymes that are difficult to express in functional forms (Figure 2). For example, functional Antarctic Candida lipase B can be generated by simply regulating the oxidation-reduction potential of a mixture of cell-free protein synthesis reactions to promote the formation of intramolecular disulfide bonds. Cellular free protein synthesis also makes it easier to introduce non natural amino acids into enzyme structures, making it particularly suitable for producing enzymes that can be selectively immobilized. For example, by introducing non natural amino acids containing chemical handles at specific locations.
Research has found that the optimal direction for immobilization can significantly improve the activity and stability of immobilized enzymes; Swartz et al. demonstrated that cell-free protein synthesis systems achieve multifunctionality of complex enzymes by expressing functional [FeFe] – hydrogenases. Moreover, the cell-free protein synthesis system is also a promising platform for producing enzymes that are toxic to recombinant hosts. For example, Lim et al. reported the successful production of phospholipase A1 using a cell-free protein synthesis system derived from Escherichia coli. Phospholipidase A1 degrades phospholipids in the cell membrane and therefore cannot be effectively produced in the cytoplasm of live E. coli cells. In the cell-free protein synthesis system, by decoupling enzyme expression from cellular physiology, a significant increase in the production of functional phospholipase A1 was achieved, which is 1000 times higher than conventional production.

Figure 2: Enhancing the production of functional enzymes by directly adding folding effectors
The complexity of cellular metabolism is another obstacle to the effective production of target chemicals, and the efficiency of most natural microorganisms is insufficient to support the high yield of target chemicals, thus meeting the current market demand. The most direct way to address these limitations is to use purified enzymes to construct cell-free metabolic pathways. The cell-free metabolic pathway based on purified enzymes can simply explain the results and optimize the participating enzymes. For example, Bujara et al. successfully demonstrated how to optimize cell-free metabolic pathways by combining them with real-time analysis methods. The cell-free enzymatic method allows for flexible design and focuses only on new pathways of target molecule production. For example, in order to eliminate the ATP driven reaction required for the conversion of glucose to pyruvate, Guterl et al. developed an artificial glycolysis pathway that only requires four enzymes. Subsequently, the enzymatic pathway for the synthesis of pyruvate was simplified by adding ethanol and isobutanol.
These results demonstrate the potential of cell-free metabolic engineering to produce industrial chemicals through artificial pathways. In addition to chemical production, Martin et al. also developed a synthetic pathway that produces 10 moles of dihydrogen by consuming 1 mole of ATP during the xylose decomposition process. In addition, the cell-free enzyme pathway has also been successfully used in bioelectricity production. Zhu et al. reported a synthetic cell-free pathway that generates nearly 24 electrons per glucose unit in an aerobic enzyme fuel cell. The energy storage density of this enzyme catalyzed fuel cell is one order of magnitude higher than that of lithium-ion batteries.
In summary, the fusion of cell-free protein technology and metabolic engineering in cell-free metabolic engineering is expected to provide an alternative pathway for the biological production of chemicals. Due to the independence of systems developed through cell-free metabolic engineering from cell viability and growth, as well as isolation from the toxicity of synthetic chemicals, they can provide greater flexibility and higher conversion efficiency.
In recent years, significant progress has been made in the design of biosynthetic pathways, construction and evaluation of metabolic modules. With the continuous improvement of cell-free protein synthesis systems, cell-free pathway prototypes have become important tools for enzyme element screening and metabolic engineering module optimization. With the continuous advancement of various technologies, the biosynthesis metabolism module and rapid prototyping technology based on cell-free protein synthesis systems will make great progress in the construction of artificial biosynthesis systems, the utilization of renewable resources to synthesize industrial chemicals, drugs, functional materials, and other high value-added compounds.
| Catalog Number | Product Name | Product Size | Applications | Price |
|---|---|---|---|---|
| GE0011 | High-Fidelity DNA Assembly Cloning Kit | 10 Reactions | Function as a DNA-guided endonuclease. | Online Inquiry |
| GE0012S | High-Fidelity DNA Assembly Master Mix | 10 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |
| GE0012L | High-Fidelity DNA Assembly Master Mix | 50 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |
| GE0012X | High-Fidelity DNA Assembly Master Mix | 250 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |